December 2015 Issue
Topics
Cotranscriptional folding: Computational modelling of self-assembly of RNA origami for universal computation
Shinnosuke Seki, Assistant Professor
Graduate School of Informatics and Engineering
University of Electro-Communications, Tokyo
Imagine a world where the basic building blocks of massive structures interacted autonomously to self-assemble into majestic landmarks such as the Eiffel Tower in Paris and the Golden Gate Bridge in San Francisco.
Needless to say the self-assembly of such huge structures is still the realm of science fiction but self-assembly is ubiquitous on the atomic scale where atoms and molecule act autonomously to form much larger structures such as liquid crystals, lipids, and layer by layer synthesis of single crystal semiconductor thin films.
Effectively exploiting self-assembly of atoms and molecules for the mass production of nano-circuits or molecular robots requires a deep understanding of the highly complex processes governing the formation of structures exemplified by self-assembled DNA-origami, as reported recently. So research on self-assembly is highly interdisciplinary with experiment-based biologists, chemists and physicists collaborating with mathematicians and computer scientists.
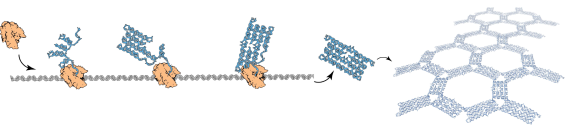
At UEC, Shinnosuke Seki is collaborating with the groups in Denmark and USA who developed so-called 'RNA origami' [1-4]. "My role is to model the self-assembly of RNA origami," says Seki. "Our research focused on a new phenomenon that we refer to as 'cotranscriptional folding'.
Here transcription refers to a process where RNA polymerase binds to a DNA sequence and produces its RNA copy sequence nucleotide by nucleotide while scanning the DNA template from one end towards the other."
Notably, unlike famous double-helical structure of DNA, RNA exists naturally as a single-strand that folds onto itself as soon as a few of its nucleotides are transcribed. This property of RNA the methodology enables the use of easy to handle single strands of RNA to construct complicated shapes.
"Using this phenomenon of cotranscriptional folding, Geary, Rothemund, and Andersen1 developed the experimental technique called RNA origami for the autonomous production of nanoscale structures," explains Seki. "As my contribution to this research, I have proposed a mathematical model of computation by cotranscriptional folding called oritatami system."
Specifically, the Oritatami system is a mathematical model of computation by cotranscriptional folding where starting from an initial 'seed' conformation, a primary building block (such as a chain of molecules) is folded cotranscriptionally over a predefined grid. "One of the important features of the Oritatami system is that it does not fold its primary structure at once," says Seki. "Instead we transcribe the fragment of the next 'delay' molecules and work dynamically from there."
This research has many applications including the realization of 'universal computation' and may enable the formation of RNA structures inside living cells.
Further information
- [1] C. Geary et.al A single-stranded architecture for cotranscriptional folding of RNA nanostructures, Science, 345 799-804 (2014).
- [2] C. Geary, P.-E. Meunier, N. Schabanel, and S. Seki, Efficient universal computation by molecular cotranscriptional folding, in preparation.
- [3] L. Kari, S. Kopecki, P.-E. Meunier, M. J. Patitz, and S. Seki, Binary pattern tile set synthesis is NP-hard, Proc. ICALP 2015, Part 1, LNCS 9134, pp. 1022-1034, (2015).
- [4] M. Ota and S. Seki, Rule set design problems for oritatami system, in revision.